|
ORGANIZATION AND EVOLUTION OF METABOLIC AND GENETIC NETWORKS |
|
|
|
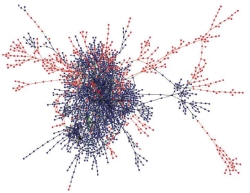
Is metabolism subject to general principles of organization? We are interested in the question of whether optimality principles are helpful in understanding the architecture and utlization of metabolic networks. Have metabolic pathways gradually evolved to perform several tasks in a maximally efficient way? We approach these questions both in simplified artificial chemistries and in the global map of known biochemical reactions across all organisms.
|
|
|
What role did metabolism play in the emergence of complex multicellular life? What
are the metabolic requirements for the
evolutionary transition from unicellular to
multicellular systems? As part of the |
|
|
Can we predict evolutionary trajectories of microbial metabolism? In
collaboration with the |
|
|
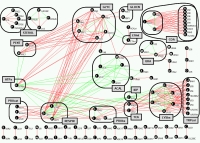
How nonlinear is metabolism? From metabolic to genetic networks How does the cell respond to genetic perturbations, and how do perturbations combine with each other to affect phenotype? We study the effects of single and double perturbations (e.g., deletions) of metabolic enzyme genes in the yeast S. cerevisiae. Deviation from expected effects in the double perturbations give rise to epistatic interactions, producing system-level functional maps of the cell |
|
|
Can we understand and reprogram metabolic regulation? One of the challenges in predicting metabolic activity at genome scale is the integration of regulatory and metabolic knowledge. We address this problem by performing high-throughput mRNA expression measurements in the bacterium Shewanella oneidensis MR-1 (whose versatile respiratory functions make it a key candidate for bioremediation and bioenergy applications), and implementing dynamic flux balance models that incorporates regulatory data and metabolite measurements. More broadly, we try to understand how metabolic pathways may be modulated to increase the production of bioenergy or useful natural products. |
|
FROM GENOME-SCALE TO ECOSYSTEM-LEVEL METABOLISM |
|
|
|
Metabolism of the human oral microbiome We have recently developed a mathematical model for the oral pathogen Porphyromonas gingivalis with the goal of understanding the role of microbial metabolism in periodontal and cardiovascular diseases caused by this microbe. We are further exploring the interaction of P. gingivalis with other oral bacteria. |
 |
Microbial community dynamics and synthetic ecology Communities of microorganisms are found nearly ubiquitously and play important roles in human health, the environment and industry. We develop mathematical models to simulate the growth of different microbial species in a community, to predict the metabolic interactions that occur between them, and to predict environments that lead to stable community structures. |